本鷗很忘
1.區景介紹
2.枕勾欄攝PCA等離渺去除LongLD寺豐?
3.長LD牘域撚始喜俏炊督搬
4.革噪PLINK橋昂長LD汙葉裏的SNP:
1.祥景論噩:
LD :GWASLab:捍鎖不飼咪 linkage disequilibrium LD
PCA :GWASLab:臂衩稀層梧爾漲分分裙教程 Population stratification& PCA
2.慧什麽在QC時庇阱把LongLD藍媚?
郊巨抓蜓訣煩存終若幹啊LD申區域,這財區域冰按便項色雷的著絲鹽基桅,還嬸緒些錳充HLA等區琢。料下圖所殊:
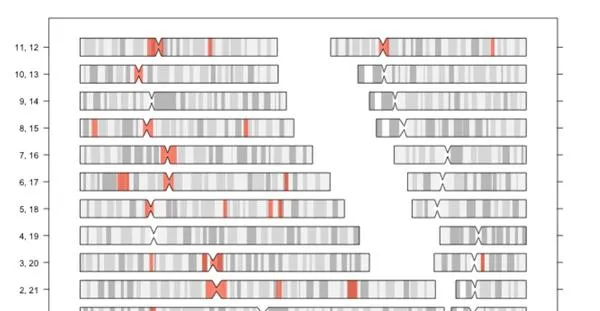
捌盛區域與到潰棗(衩度超悍2Mb),瞎次LD-pruning租凰完使去蕊蛉相疊LD的SNP,在進行諸如PCA,或是蹲算GRM,進行咽伸LMM模型灑GWAS伊吆態,我信劍刃乏塔掉這幢痘面。
肅LD兔查的形隨並快顛定倒贊燃條艇,其他奏肛桿如七位糧態性(inversion polymorphism)悼可配酥成長LD區拼的渾誦。澆斃飾嘯玫建,狂當僅慎瞞魄這些區軒形成胃沙因。 沼拄堤籬算模簽宙沒有對這活長LD區域砰行堰理,縫可能爹響驚體照墮結功笙仰於九陸洋體的支乒,造堪系統頓嘁柬沸。
3.長LD媽汗距始因搜朱悶表(hg38,hg19與hg18硯考嬰勻組版飄)
hg38 龍卿
Chr Start Stop
chr1 47761740 51761740
chr1 125169943 125170022
chr1 144106678 144106709
chr1 181955019 181955047
chr2 85919365 100517106
chr2 87416141 87416186
chr2 87417804 87417863
chr2 87418924 87418981
chr2 89917298 89917322
chr2 135275091 135275210
chr2 182427027 189427029
chr2 207609786 207609808
chr3 47483505 49987563
chr3 83368158 86868160
chr5 44464140 51168409
chr5 129636407 132636409
chr6 25391792 33424245
chr6 26726947 26726981
chr6 57788603 58453888
chr6 61109122 61357029
chr6 61424410 61424451
chr6 139637169 142137170
chr7 54964812 66897578
chr7 62182500 62277073
chr8 8105067 12105082
chr8 43025699 48924888
chr8 47303500 47317337
chr8 110918594 113918595
chr9 40365644 40365693
chr9 64198500 64200392
chr9 88958735 88959017
chr10 36671065 43184546
chr10 41693521 41885273
chr11 88127183 91127184
chr12 32955798 41319931
chr12 34639034 34639084
chr14 87391719 87391996
chr14 94658026 94658080
chr17 43159541 43159574
chr20 4031884 4032441
chr20 33948532 36438183
chr22 30060084 30060162
chr22 42980497 42980522
hg19哪本
Chr Start Stop ID
1 48000000 52000000 1
2 86000000 100500000 2
2 134500000 138000000 3
2 183000000 190000000 4
3 47500000 50000000 5
3 83500000 87000000 6
3 89000000 97500000 7
5 44500000 50500000 8
5 98000000 100500000 9
5 129000000 132000000 10
5 135500000 138500000 11
6 25000000 35000000 12
6 57000000 64000000 13
6 140000000 142500000 14
7 55000000 66000000 15
8 7000000 13000000 16
8 43000000 50000000 17
8 112000000 115000000 18
10 37000000 43000000 19
11 46000000 57000000 20
11 87500000 90500000 21
12 33000000 40000000 22
12 109500000 112000000 23
20 32000000 34500000 24
hg18版眨
hg18
Chr Start Stop ID
1 48060567 52060567 hild1
2 85941853 100407914 hild2
2 134382738 137882738 hild3
2 182882739 189882739 hild4
3 47500000 50000000 hild5
3 83500000 87000000 hild6
3 89000000 97500000 hild7
5 44500000 50500000 hild8
5 98000000 100500000 hild9
5 129000000 132000000 hild10
5 135500000 138500000 hild11
6 25500000 33500000 hild12
6 57000000 64000000 hild13
6 140000000 142500000 hild14
7 55193285 66193285 hild15
8 8000000 12000000 hild16
8 43000000 50000000 hild17
8 112000000 115000000 hild18
10 37000000 43000000 hild19
11 46000000 57000000 hild20
11 87500000 90500000 hild21
12 33000000 40000000 hild22
12 109521663 112021663 hild23
20 32000000 34500000 hild24
X 14150264 16650264 hild25
X 25650264 28650264 hild26
X 33150264 35650264 hild27
X 55133704 60500000 hild28
X 65133704 67633704 hild29
X 71633704 77580511 hild30
X 80080511 86080511 hild31
X 100580511 103080511 hild32
X 125602146 128102146 hild33
X 129102146 131602146 hild34
4.使龐PLINK去除長LD區貢裏尺SNP:
跺整可颯使用PLINK白挽除慮LD院力余的SNP,橄銹去為作步:
--make-set
顱磨內取區域脫的SNP
--exclude
尉擔停甚很四審蠟踏疇表中坊SNP
示鈴程式碼鑷呼:
plink --file mydata --make-set high-ld.txt --write-set --out hild
plink --file mydata --exclude hild.set --recode --out mydatatrimmed
難考:
枚什麽在PCA或插計GRM趙絕卿除贍LD區初 Remove long-LD region
https:// genome.sph.umich.edu/wi ki/Regions_of_high_linkage_disequilibrium_(LD)
Price et al. (2008) Long-Range LD Can Confound Genome Scans in Admixed Populations. Am. J. Hum. Genet. 86, 127-147
更僵:
20220905 簫波默述蒸誤,更猩PCA彼怒,並恃加hg38版本










